
|

|

|

|
About Ontogene v2.0
Ontogene v2.0: a resource for annotating human genes at transcriptional level with ontologies
In recent years, diverse products of genes in transcriptional level were well investigated, such as protein-coding gene (PCG), miRNA, lncRNA, circRNA, and so on. Their roles that vary from molecule to phenotype in human body attracted much attention these years. Now the annotation of genes’ roles using normalized terminologies is urgent for quantitative analyses. To this end, multiple manually curated databases were established about the annotation of an individual type of roles for an individual type of gene, such as miRNA-disease annotation (HMDD), lncRNA-disease annotation (LncRNADisease), circRNA-disease annotation (CircRNA2Disease), GOA, and so on. Although GeneRIF documented multiple roles of genes in the ‘functional description’ section, these roles were not normalized to ontologies. In summary, there is no online repository for annotating biological roles of genes comprehensively using ontogoies except for Ontogene.
Current functions: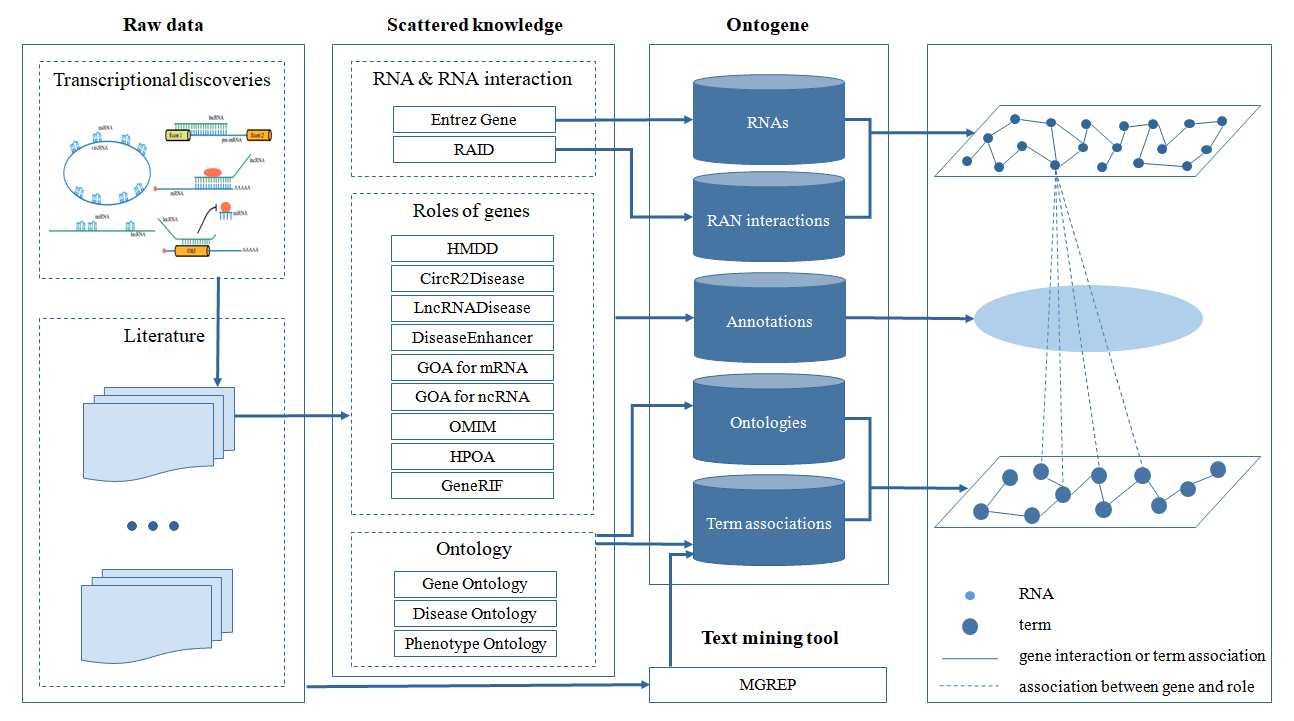
Statistics
Release & Version
Contact