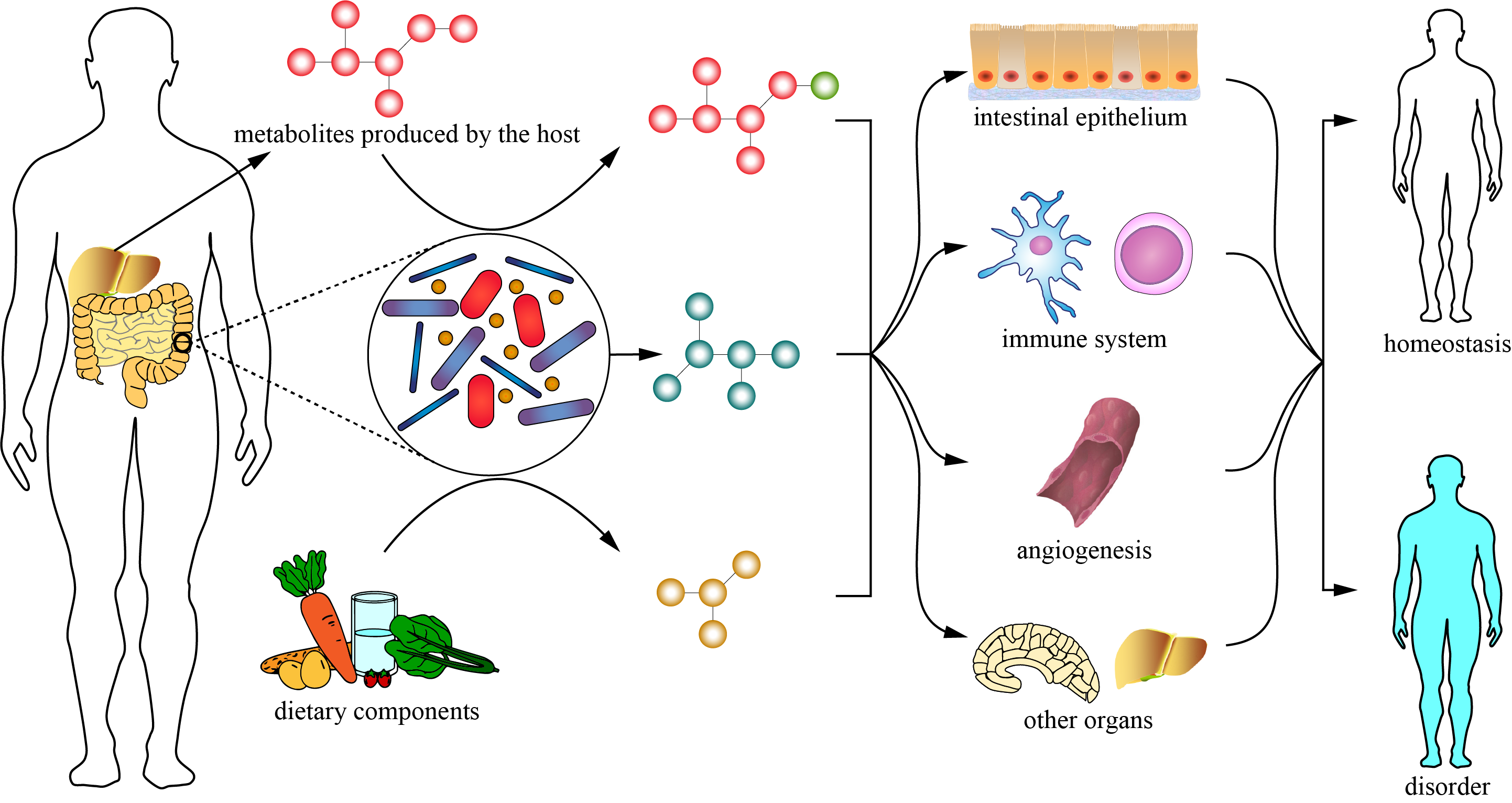
About gutMGene v1.0
gutMGene: a comprehensive database for target genes of gut microbes and microbial metabolites
Metagenomic sequencing of fecal samples has identified 3.3×106 non-redundant microbial genes from up to 1,500 different species. One of the contributions of gut microbiota to host biology is the circulating pool of bacterially derived small-molecule metabolites. It has been estimated that 10% of metabolites found in mammalian blood are derived from the gut microbiota, where they can produce systemic effects on the host through activating or inhibiting gene expression. Currently, detailed information on target genes of gut microbes and microbial metabolites is scattered in literatures and no online repository is available for collecting these data.
Here, we manually extracted microbe-metabolite, microbe-target, and metabolite-target relationships in Human and Mouse from almost 400 publications. All of these relationships were experimentally validated in vivo or in vitro and measured by RT-qPCR, high performance liquid chromatography, 16S rRNA sequence, and so on.
Currently, users can
• Download all manually curated relationships.
• Achieve relationships by browsing or searching.
• Submit novel relationships to the gutMGene database.
Release & Version
Jun 20, 2021, the gutMGene v1.0 was released.
Contact
Liang Cheng, Ph. D.
Chief Editor of Current Gene Therapy
Professor and PI of Harbin Medical University
Mail: liangcheng@hrbmu.edu.cn
Statistics
Totally, 1,331 relationships between gut microbes, microbial metabolites, and genes in Human were manually extracted.
| Type | Number | |
|---|---|---|
| Gut microbe | 332 | |
| Metabolite | 207 | |
| Gene | 223 | |
| Relationship | microbe-metabolite | 724 |
| microbe-target | 298 | |
| metabolite-target | 309 | |
Totally, 2,349 relationships between gut microbes, microbial metabolites, and genes in Mouse were manually extracted.
| Type | Number | |
|---|---|---|
| Gut microbe | 209 | |
| Metabolite | 149 | |
| Gene | 544 | |
| Relationship | microbe-metabolite | 776 |
| microbe-target | 744 | |
| metabolite-target | 829 | |