
|

|

|

|
Overview
Here, we provided the tutorial of Ontogene v2.0. It includes the following aspects.
1. Tutorial for search page
2. Tutorial for annotation table
3. Tutorial for gene-term-term-gene path visualization
4. Tutorial for term-gene-gene-term path visualization
5. Introduction for our sister projects
6. Question and answer
The search page allows users to input a term of ontology or a gene for querying annotations of genes as following.
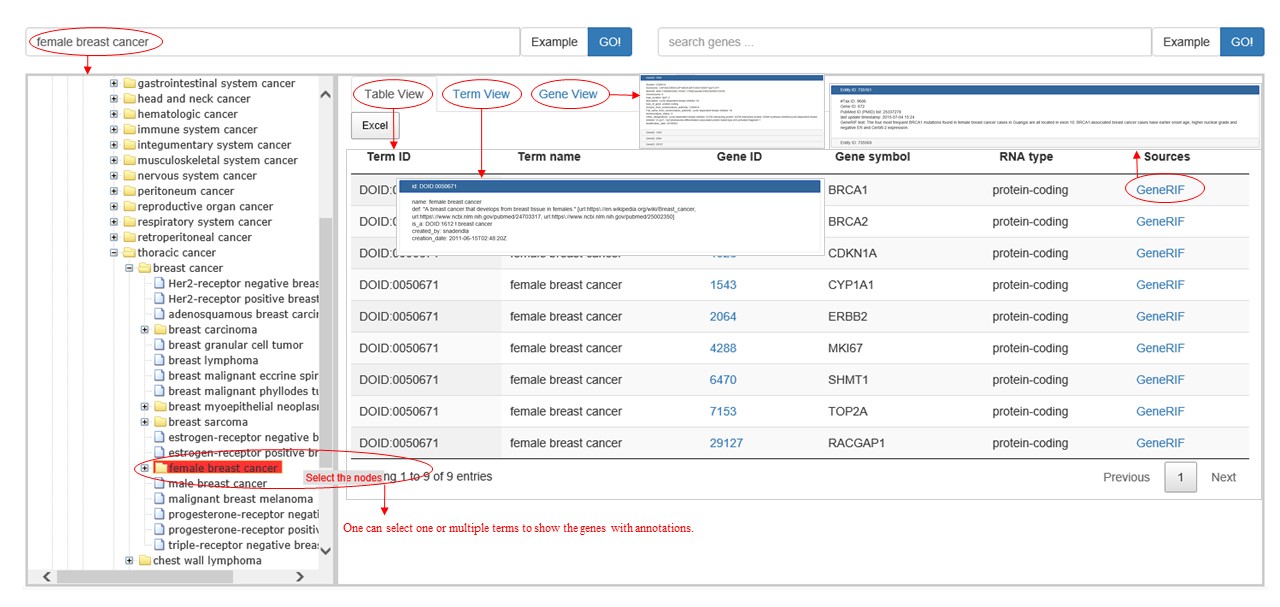
After searching a term of ontology, the tree structure of this term is shown in the left, and its genes are shown in the right in a 'Table View'. Each line in the table contains an association between a gene and a term, which involves term ID, term name, gene symbol, gene ID, the type of gene, and sources. The details of the description could be shown by clicking the data source link. In addition, 'Term View' and 'Gene View' show the details of the terms and genes in the 'Table View'.
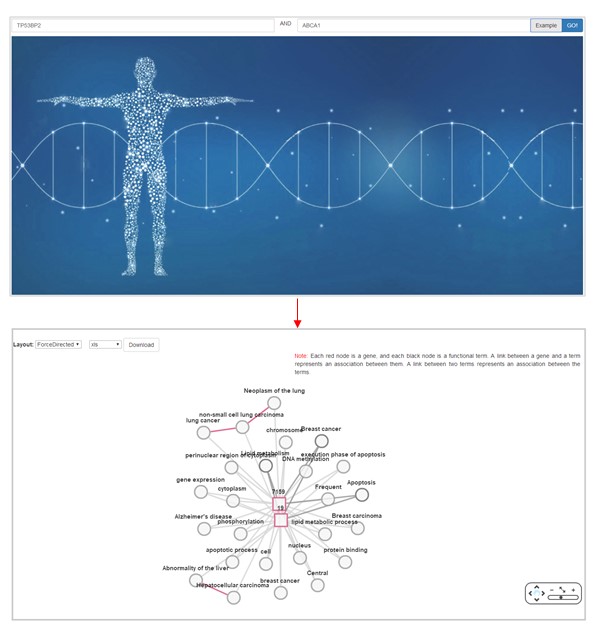
Two genes can be linked by their common functions, or by the associations between their terms in Ontogene v2.0 as following.
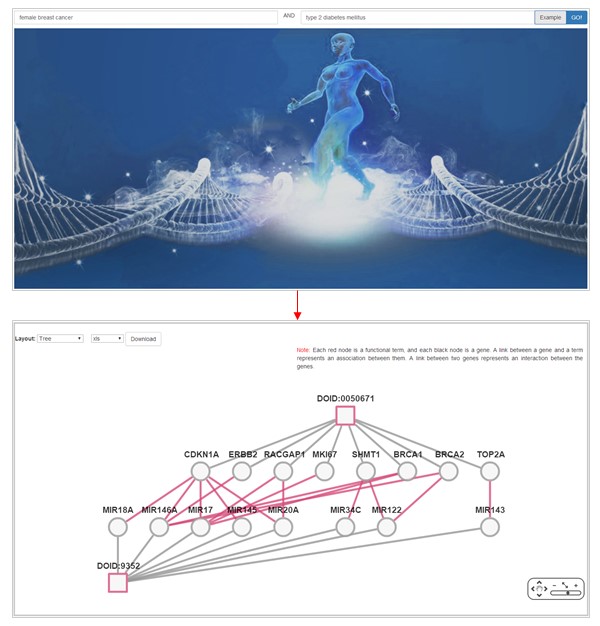
Two terms can be linked by their common genes, or by the interactions between their genes in Ontogene v2.0 as following.
All of our previous researches focused on the human disease system biology, such as exploring the casual, biomarker of human disease in molecular and phenotype level. Three sister projects have been conducted by us.
6.1 How to download the database?
Here, we provide the page to download all the data from the web.
We also provide web services for users to access our data.
6.2 How to show users' feedback?